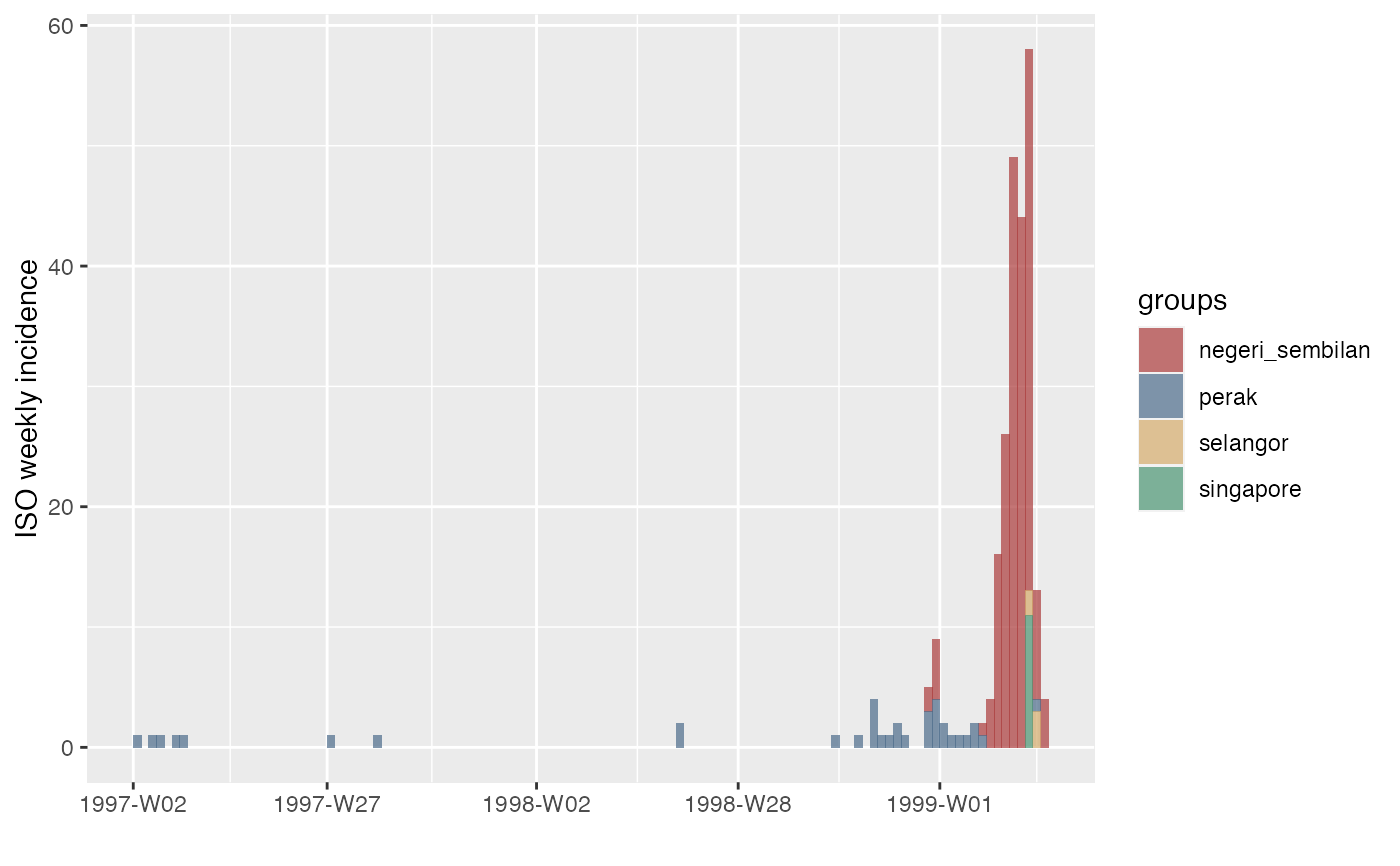
These data describe incidence of human cases of Nipah virus encephalitis in Malaysia and Singapore from January 1997 through April 1999.
nipah_malaysia
Format
A data frame with 49 rows and 5 columns
- date
Onset date (weekly)
- perak
Number of cases (Perak State, Malaysia)
- negeri_sembilan
Number of cases (Negeri Sembilan State, Malaysia)
- selangor
Number of cases (Selangor State, Malaysia)
- singapore
Number of cases (Singapore)
Source
Pulliam et al. (2011)
References
J.R.C. Pulliam, et al. 2011. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. _Journal of the Royal Society Interface_, 9(66), 20110223. https://doi.org/10.1098/rsif.2011.0223
Author
Data from Funk et al. (2016), provided by Juliet Pulliam (github.com/jrcpulliam).
Examples
#> # A tibble: 6 × 5 #> date perak negeri_sembilan selangor singapore #> <date> <int> <int> <int> <int> #> 1 1997-01-04 0 0 0 0 #> 2 1997-01-11 1 0 0 0 #> 3 1997-01-18 0 0 0 0 #> 4 1997-01-25 1 0 0 0 #> 5 1997-02-01 1 0 0 0 #> 6 1997-02-08 0 0 0 0## convert data to incidence object and plot epicurve using the incidence package library(incidence) cases <- subset(nipah_malaysia, select = c("perak", "negeri_sembilan", "selangor", "singapore")) i <- as.incidence(cases, dates = nipah_malaysia$date, interval = 7L) plot(i)